User Manual
Written by: Piotr Klukowski
Published: July 29, 2024, 10:03 a.m.
Axes disambiguation
This property specifies the order of axes in the uploaded spectrum file. Its possible values are stored in "Project Storage" as multiple choice list, such as the one presented in the figure below.
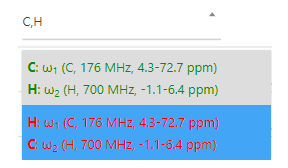
Each option in the list presents a possible mapping of axes specific to the selected experiment type (e.g. 13C-HSQC) to dimensions in the uploaded spectrum. Options printed in green are more likely to be correct than the red options. Each option contains a list of axes in the following format:
A: B (C, D MHz, E ppm)B - axis ID (ω1, ω2, ...)
C - axis name extracted from the header of the uploaded spectrum file
D - axis range in MHz extracted from the header of the uploaded spectrum file
E - chemical shift range in ppm extracted from the header of the uploaded spectrum file
A common mistake is to wrongly disambiguate 1H axes in 3D/4D NOESY and HCCH-TOCSY experiments. Some difficulties in spectrum disambiguation might be resolved by inspecting spectrum visualizations available in "Project Storage → Info", and comparing them with the examples presented in the table below.
| Spectrum type | Axis labels | Example visualizations |
| 13C-resolved [1H-1H] NOESY | C HC - hydrogen covalently bonded to C H - hydrogen interacting through-space with HC-C |
HC-H, HC-C, H-C |
| 15N-resolved [1H-1H] NOESY | N HN - hydrogen covalently bonded to N H - hydrogen interacting through-space with HN-N |
HN-H, HN-N, H-N |
| CCH-TOCSY | C1 - carbon 1 C2 - carbon 2 H1 - hydrogen covalently bonded to C1 |
C2-C1, H1-C1, H1-C2 |
| HBHAcoNH | N HN - hydrogen covalently bonded to N H |
HN-H, HN-N, H-N |
| HCCH-TOCSY / HCCH-COSY | C HC - hydrogen covalently bonded to C H |
H-HC, HC-C, H-C |
| HCcoNH | N HN - hydrogen covalently bonded to N H |
HN-H, HN-N, H-N |
| 15N-TOCSY | N HN - hydrogen covalently bonded to N H |
H-N, HN-N, H-HN |
Spectrum unfolding
This property specifies the ID of the axis with folded/aliased signal. If aliased signals have opposite amplitude to regular (unaliased) signals, one can limit the scope of the automated unaliasing procedure to positive or negative signals only.
For example in 13C-resolved [1H, 1H]–NOESY it is common to fold signals along 13C dimension. Assuming that 13C correspond to the second dimension in the uploaded spectrum, option ω2 should be selected.
Spectrum tags
It is possible to provide additional information about the uploaded spectrum using a subset of predefined tags:
| Tag | Description |
| @ALI | Only signals from aliphatic atoms have been recorded |
| @ARO | Only signals from aromatic atoms have been recorded |
| @NEG | Analyze only signals with negative signal amplitude |
| @POS | Analyze only signals with positive signal amplitude |
| wA:B | Shift spectrum along axis A by B ppm (e.g w1:0.4) |
| H:B, HN:B, HC:B, H1:B, H2:B | Only atoms defined by CYANA selection B are observable in dimension H/HN/HC/H1/H2 (e.g. HC:ARO, HN:100-150) |
| important | Forces NOESY peaks to be assigned whenever possible in the automated NOE assignment. Use this tag only for carefully curated NOESY peak lists that do not contain any artifacts. Equivalend to using assign_noartifact in CYANA. |
All tags associated with a spectrum can be provided in the "Project Storage" as comma-separated values. For example, signals in a spectrum with the tags:
@ALI,w1:0.4Spectra name convention and supported data formats
NMRtist uses Sparky (*.ucsf) format to store and process NMR data. Spectra uploaded in other formats are converted automatically to Sparky format during the upload process, as presented in the table below.
| Format | Required file extension | Description |
| Sparky | ucsf | NMRtist native format, no conversion required |
| NMR pipe | pipe | File is automatically converted using the pipe2ucsf tool available in Sparky/bin directory |
| Bruker | zip | Bruker spectra are uploaded as single zip file, which contains the full Bruker directory tree of the spectrum. Processed spectrum data (i.e. 2rr or 3rrr) must be available in the pdata subdirectory. The uploaded zip file is automatically extracted and converted using the bruk2ucsf tool available in Sparky/bin directory. An example zip file with a Bruker spectrum can be downloaded here. |
| XEASY | zip | Spectra in XEASY format are uploaded as single zip file, which contains 2 files (*.3D.16, *.3D.param). The uploaded zip file is automatically extracted and converted using Yokochi converters. An example zip file with a XEASY spectrum can be downloaded from here. |
In case of problems with automated online conversion, it is recommended to convert spectra to Sparky format locally and then proceed with the upload.
If the filename of an uploaded spectrum matches the pattern
spectrumType_spectrumTagsfor example
C13NOESY_@ALI@NEGthen the spectrum type and tags are set automatically in the "Project Storage". Only @ALI, @ARO, @POS and @NEG tags are allowed in the filename.
Other data file formats
Starting from 27.03.2023, in addition to NMR spectra, NMRtist supports the following file formats: (a) manual peak lists (*.list, *.peaks), (b) chemical shift lists (*.prot), (c) chemical shift statistics (*.stats), (c) lower/upper distance restraints (*.lol/*.upl), (d) Talos angle restraints (*.aco), and (e) protein structure (*.pdb). These files can be uploaded to the project storage and used as inputs for the applications. The table below contains example files in formats compatible with NMRtist.
| File type | Format | Example files | Comment | |
| Manual peak list | XEASY (*.peaks) Sparky (*.list) CCPN (*.nef) |
N15HSQC (Sparky, unassigned) N15HSQC (Sparky, assigned) N15NOESY (Sparky, unassigned) N15NOESY (Sparky, assigned) N15HSQC (XEASY, unassigned) N15HSQC (XEASY, assigned) N15NOESY (XEASY, unassigned) N15NOESY (XEASY, assigned) C13HSQC (NEF, assigned) |
In the uploaded peak list, you may specify both assigned and unassigned peaks. Please note that NMRtist disregards any peak assignments, allowing you to manually add them to your project using *.prot files. For all application types, you can utilize manual peak lists as input data. | |
| Chemical shift lists | CYANA (*.prot) | Download | The chemical shift list, which serves as input for the application, fixes the chemical shifts to the specified values in the uploaded file. The application accepts both complete and partial chemical shift lists, accommodating various data availability. | |
| Chemical shift statistics | CYANA (*.stats) | Download |
The chemical shift statistics file functions similarly to the chemical shift list file (.prot) but applies softer constraints. Instead of fixing the chemical shift to predefined values (.prot), NMRtist prioritizes the shifts specified in the *.stat file, assigning them only when feasible. Each specified chemical shift in the stats file must have a non-zero "tolerance" value. This tolerance helps the method identify favorable regions in the spectra where the chemical shift is anticipated. Chemical shift statistics files are useful for chemical shift transfer applications. In this scenario, the target protein spectra are uploaded to the project, while the homolog protein chemical shifts are stored in a *.stat file. | |
| Lower/upper limit restraints | CYANA (*.lol, *.upl) | Download | During the CYANA structure calculation step, specified restraints are employed alongside the restraints extracted from NMR spectra to enhance the accuracy of the resulting model. | |
| TALOS angle restraints | TALOS (*.aco) | Download | In the ARTINA workflow, specified restraints take the place of the TALOS step. The uploaded aco file will be used directly during the structure calculation phase. | |
| Protein structure | PDB (*.pdb) | Download | Protein Structure Model (AlphaFold prediction, X-ray, or homolog structure):
Enhances the accuracy of chemical shift assignment (if the uploaded model is within <3Å backbone RMSD of the measured protein)
Can be utilized for structure-based assignment.
Remark I: We recommend using structure files solely for chemical shift assignment. For structure determination, the structure file can serve as input; however, the results will be heavily biased towards the input structure. Remark II: Uploading protein bundles is preferable to single-model PDB files (e.g., X-ray). AlphaFold typically generates five structure proposals. While it is possible to use a single (best) proposal as input, we advise creating a PDB file containing all five models before uploading it to NMRtist projects. |
The files stored in the above table are compatible with the following protein sequence:
MKIISISETPNHNTMKITLSESREGMTSDTYTKVDDSQPAFINDILKVEGVKSIFHVMDFISVDKENDANWETVLPKVEAVFELEHHHHHH
Application call status
| Status | Description | Allowed user actions |
| Waiting | Application call request has been registered in the NMRtist system. Waiting for computational resources. | OpenCancelRemove |
| Running | Application is currently running on NMRtist computational node (or computer cluster) | OpenCancelRemove |
| Finished | Application results have been fetched from NMRtist computational node (or computer cluster) | OpenCancelRemove |