About NMRtist
NMRtist is a cloud computing service for the fully automated analysis of protein NMR spectra (e.g. peak picking, chemical shift assignment, structure determination) using deep learning-based approaches. Each project created in NMRtist receives 30 GB of private storage, which can be filled by experimental data and analyzed using the available applications. You don't need to have any hardware resources or follow complex software configuration processes. NMRtist applications can be executed by just few mouse clicks in your web browser. All calculations are executed on NMRtist computational nodes, making the results available for download from NMRtist website.
ARTINA is a deep learning-based application for end-to-end protein structure determination by NMR spectroscopy. Using as input NMR spectra and the protein sequence, the method identifies automatically (strictly without any human intervention): cross-peak positions, chemical shift assignments, upper limit distance restraints, and the protein structure. ARTINA deep learning models have been trained with over 600 000 cross-peak examples from more than 1300 2D-4D spectra. The method demonstrated its ability to solve structures with a median backbone RMSD of 1.44 Å to PDB reference, and identified correctly 91.36% of the chemical shift assignments. View our short video tutorial to learn how to get started with ARTINA.
ARTINA and NMRtist can automate tasks such as cross-peak detection in 2D-4D NMR spectra, de novo chemical shift assignment of protein monomers and protein-ligand complexes, de novo structure calculation, structure-based chemical shift assignment, and chemical shift transfer. A more comprehensive list of system use cases can be found in the Articles & Tutorials section [link].
You can use the NMRtist platform free-of-charge (academic users) to perform automated peak picking, shift assignment, or full structure determination. Create a free account to use all functions of the service, or start an anonymous project by pressing the button below.
Recommended articles
Video Tutorial
This video tutorial introduces beginners to the NMRtist system, guiding them through the process of submitting an automated protein structure determination job, and showcasing representative results from such a job.
Our NMR Data Analysis Workflow
Artificial Intelligence for NMR Applications (ARTINA) is a deep learning-based approach to fully automated NMR protein structure determination. The method takes as input only NMR spectra and the protein sequence, and delivers automatically: peak lists, shift assignments, distance restraints, and the structure.
NMRtist Use Cases
This article summarizes the most common use cases of NMRtist and ARTINA, such as structure-based chemical shift assignment, chemical shift transfer, or de novo protein structure determination.
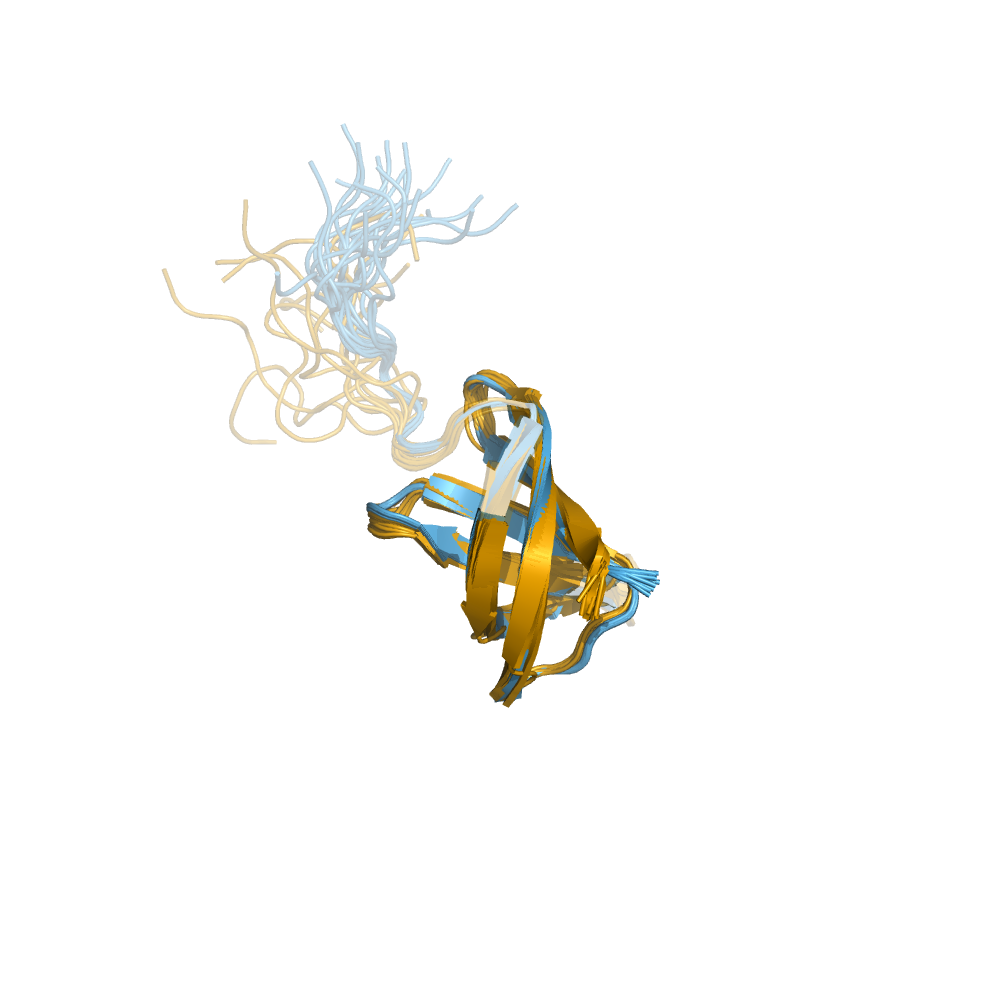
Examples of automatically determined structures

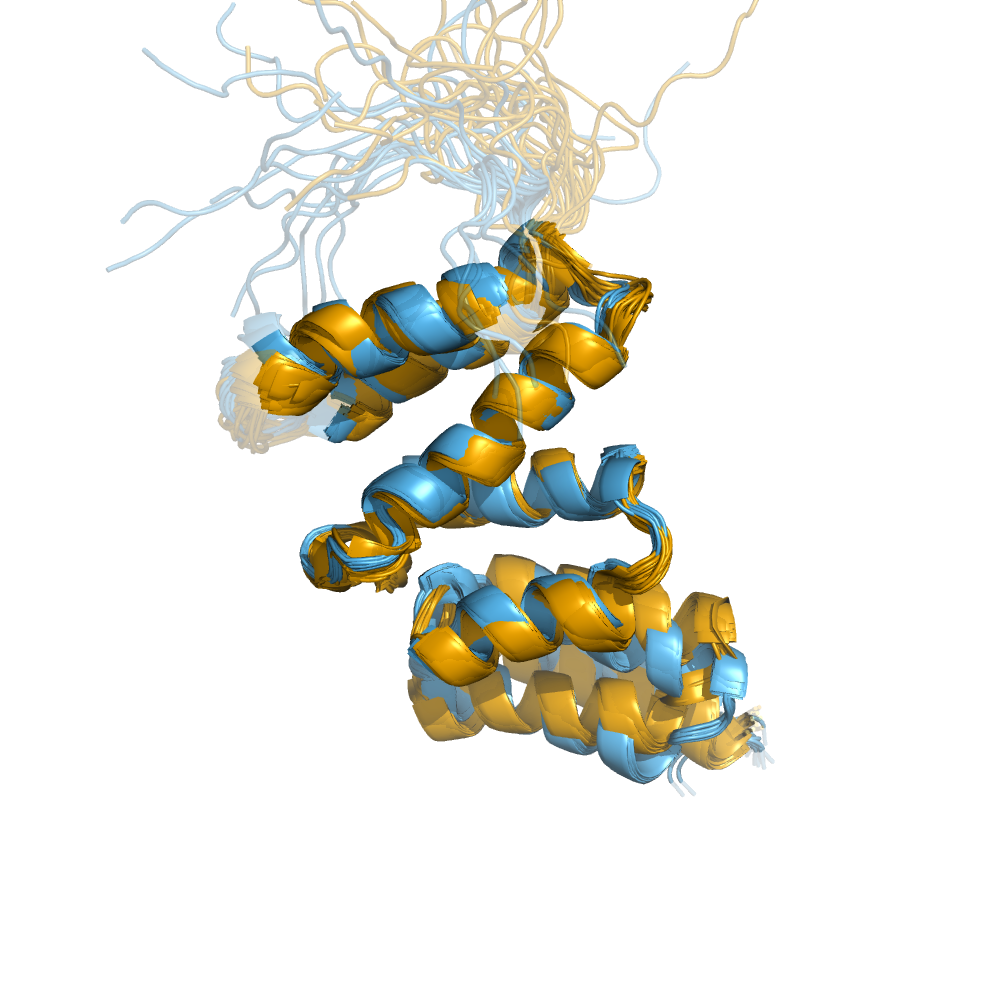
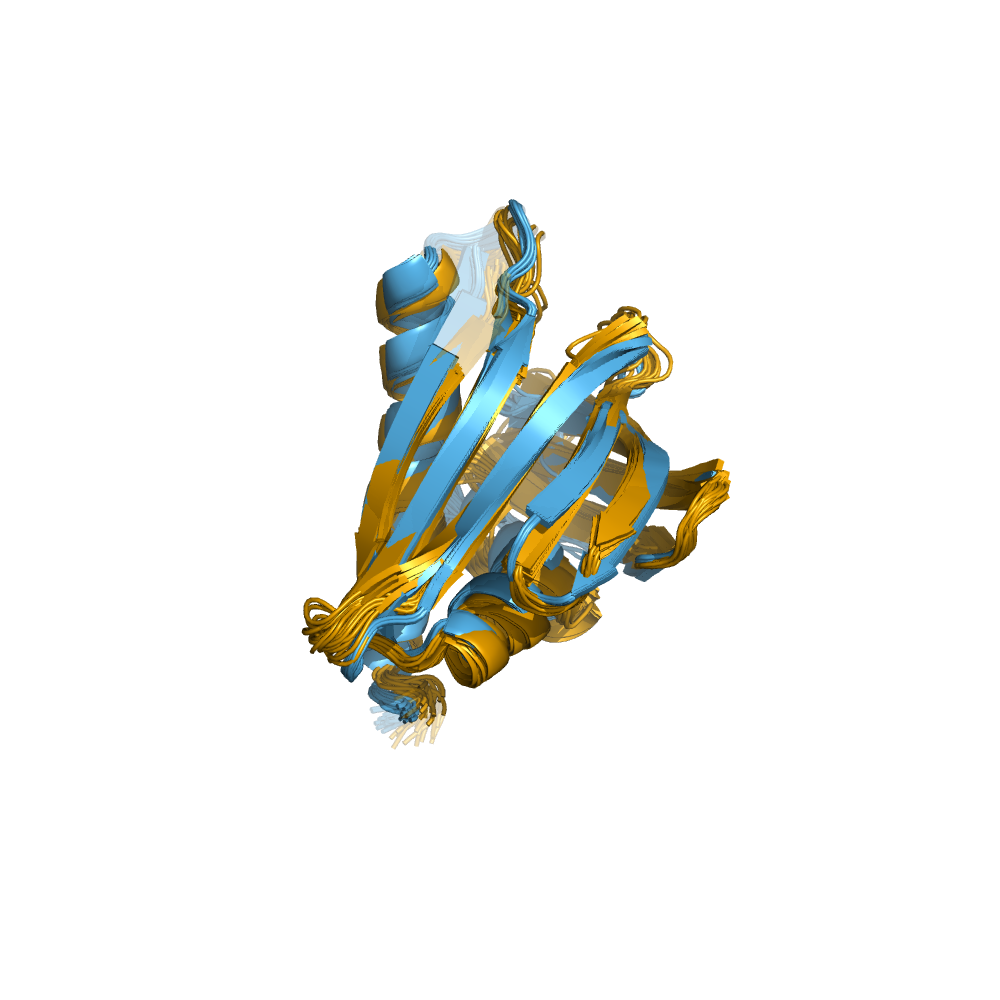
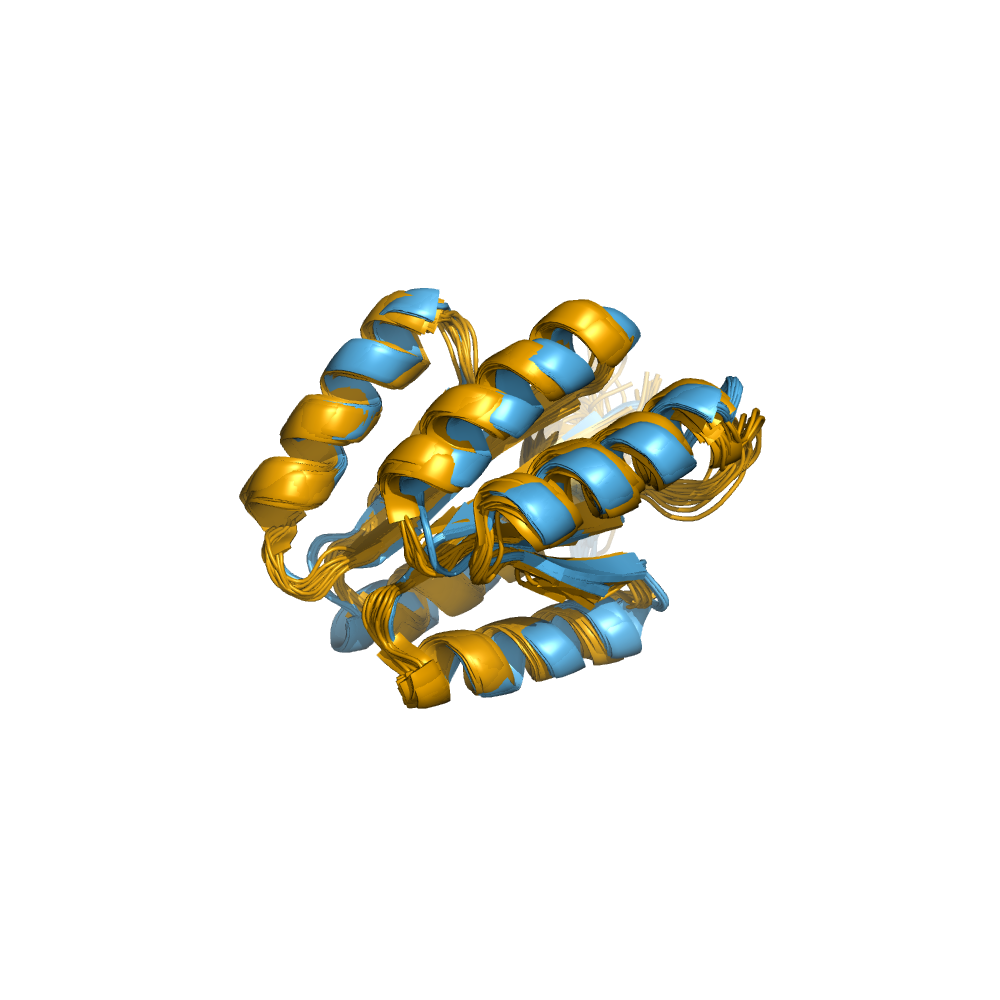
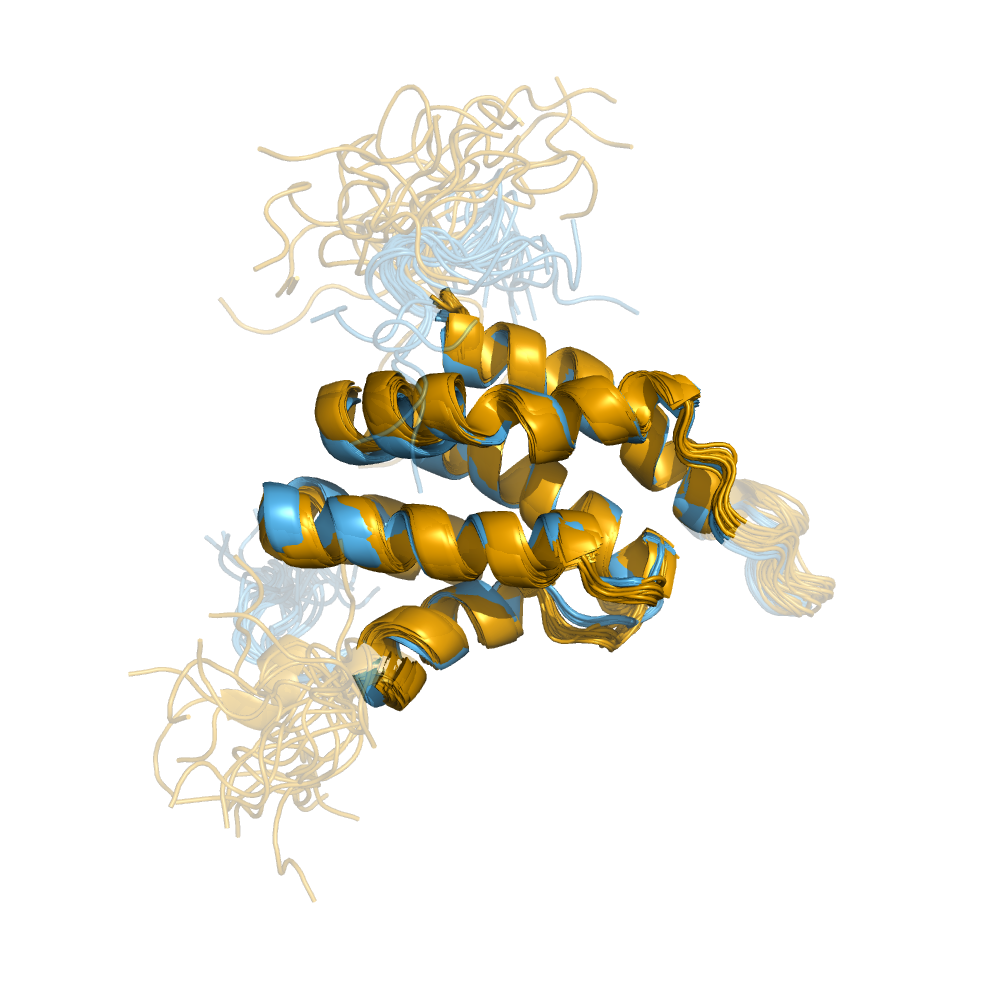


News
July 31, 2024, 11:01 a.m.
New Release: custom protein systems and enhanced data validation
In the new NMRtist release, our developments were aimed at facilitating a broader range of data analysis tasks in macromolecular NMR spectroscopy. In addition to existing features, you can now define custom protein systems while creating new projects, which includes, among others, custom residue types and complexes of proteins with small molecules, peptides, and metal ions. In principle, the new version may be used to facilitate other types of macromolecules, such as RNA and DNA, but our algorithms and deep learning models have been trained and tested explicitly for protein data.
The new version also brings updates that provide new insights into NMR data and make the use of the platform more convenient. For example, the chemical shift assignment now presents a visualization of “control points” that helps resolve issues with spectra referencing – one of the common hurdles NMRtist users faced in the previous version of the system.
We are currently working to enable methyl assignment and further support for solid-state experiments on our platform.
Jan. 15, 2024, 8:14 p.m.
[Manuscript] The 100-protein NMR spectra dataset: A resource for biomolecular NMR data analysis
Open dataset containing 1329 2D-4D NMR spectra that allow the reproduction of 100 protein structures from original measurements. This dataset was originally compiled for the development of the ARTINA deep learning-based spectra analysis method (see https://nmrdb.ethz.ch and the manuscript).
Nov. 30, 2023, 8:15 p.m.
[Manuscript] Time-optimized protein NMR assignment with an integrative deep learning approach using AlphaFold and chemical shift prediction
Our new study, recently accepted in Science Advances (https://www.science.org/doi/full/10.1126/sciadv.adi9323), explores the integration of in-silico predictions like AlphaFold with ARTINA, enhancing the efficiency and accuracy of NMR data analysis. This research represents a significant leap towards data-efficient use of our system for protein studies.
Feb. 2, 2023, 8:39 p.m.
[Manuscript] NMRtist: an online platform for automated biomolecular NMR spectra analysis
Our manuscript (application note), presenting the NMRtist platform, has been accepted for publication in Bioinformatics (https://doi.org/10.1093/bioinformatics/btad066).
Dec. 21, 2022, midnight
NMRtist usage
Since the release of the platform in February 2022, NMRtist analysed 4 368 2D/3D/4D NMR spectra, completed 1 100 automated chemical shift assignment and 444 automated structure determination jobs.
Dec. 20, 2022, midnight
ARTINA and NMRtist presented to the broader audience
Between 06.2022 and 01.2023, we presented ARTINA and NMRtist at several NMR events, including: Chianti Workshop (Principina Terra, Italy), EUROMAR (Utrecht, The Netherlands), EMBO Practical Course (Basel, Switzerland), EMBO Lecture Course (Berhampur, India), Biomolecular NMR: Advanced Tools, Machine Learning (Gothenburg, Sweden), and ICMRBS (Boston, USA).
Oct. 19, 2022, midnight
[Manuscript] Rapid protein assignments and structures from raw NMR spectra with the deep learning technique ARTINA
Our manuscript, presenting the ARTINA workflow for rapid assignment and structure determination, has been published in Nature Communications (https://doi.org/10.1038/s41467-022-33879-5).
Oct. 2, 2021, midnight
Biomolecular NMR: Advanced Tools workshop
NMRtist was presented at the Biomolecular NMR: Advanced Tools workshop (29.09-01.10 2021). All participants of the training, supervised by Prof. Peter Güntert and Dr. Piotr Klukowski, submitted datasets to the platform, obtaining automatically determined structures and/or assignments.